Pandas is a really powerful data analysis library in Python. This is not an exhaustive
reference to its methods. I did my best to include much of the most used and
more difficult to remember methods I have used over the last few years.
I encourage anyone attempting to work through this page to first get a solid grasp of the
Python language generally. This will make understanding the Pandas methods much easier to
comprehend.
Here I mostly cover accessing and handling data. The Pandas library is rather large and I
would encourage anyone reading this to also reference the documentation availible on Pandas
API here.
I use more applied methods in the data project examples pages. But understanding the methods to
access and handle data are an essential first step.
Basics of a DataFrame
Let's first go over what a dataframe is and its parts. A dataframe is the most common way to display and subsequently analyze data stored in xyz form.
Basic dataframe parts:
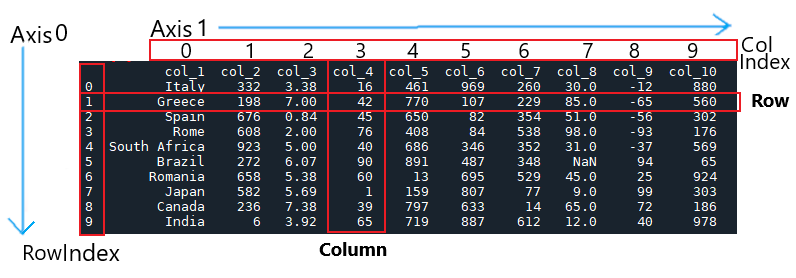
Columns and rows are obvious. What may seem more complicated is the index—which technically is not
a column. The index is used to organize data at specific positions. It is like a grid coordinate
system to organizing data—essentially this is like an array.
Many of the methods on this page may seem abstract. Notably the indexing methods. But once you know
your way around a dataframe many of the operations pertaining to analyzing a dataframe will be more
easily understood. Once you can access the data, you can apply the necessary operations to it.
The examples here include dataframes that are rather small to illustrate methods. In all practicality dataframes
are usually anywhere from hundreds, thousands to millions of data points in size.
Simple Imports and Exports
To import or export most data in pandas you can do so with the following:
Import/export data and warnings when importing in Pandas:
import pandas as pd
import numpy as np
df = pd.read_<FileType>(r'<filePath>')
#Read file by Type:
pd.read_csv(r'filePath', engine='python')
pd.read_json(r'filePath')
pd.read_excel(r'filePath')
pd.read_hdf(r'filePath')
pd.read_sql(r'filePath')
pd.read_fwf(r'filePath') #for txt docs
#On occasion you run into a utf-- codec error;
#In most cases, to circumvent this error add engine='python' argument
pd.read_excel(r'filePath', engine='python') #reads through UnicodeDecodeError: 'utf-8' codec can't...
#Save/Export by file by type:
df.to_csv(r'filePath')
df.to_json(r'filePath')
df.to_excel(r'filePath')
df.to_hdf(r'filePath')
df.to_sql(r'filePath')
#Turn off all warnings:
import warnings
warnings.simplefilter(action='ignore', category=FutureWarning)
Basic data import example:
"""
Import some data. The variable (df_0) is assigned to the dataframe.
The (r) preceding the file path tells Python to read the path as a raw string.
If you do not use it it may return an exception because it may interpret the
backslashes as escape characters.
"""
In[]: df_0 = pd.read_csv(r'path\to\file\some_file_name.csv')
"""
Look at the dataframe by calling df_0. This data is completely random and
has no significance except to demonstrate some initial methods here. I will
use this dataframe and others in subsequent examples.
"""
In[]: df_0
Out[]:
col_1 col_2 col_3 col_4 col_5 col_6 col_7 col_8 col_9 col_10
0 Italy 332 3.38 16 461 969 260 30.0 -12 True
1 Greece 198 7.00 42 770 107 229 85.0 -65 False
2 Spain 676 0.84 45 650 82 354 51.0 -56 False
3 Rome 608 2.00 76 408 84 538 98.0 -93 True
4 South Africa 923 5.00 40 686 346 352 31.0 -37 True
5 Brazil 272 6.07 90 891 487 348 NaN 94 True
6 Romania 658 5.38 60 13 695 529 45.0 25 False
7 Japan 582 5.69 1 159 807 77 9.0 99 False
8 Canada 236 7.38 39 797 633 14 65.0 72 False
9 India 6 3.92 65 719 887 612 12.0 40 True
Some highlighted imports:
'''
Here are some imports that you may use in the future.
I will use these in some data project examples.
'''
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import sklearn as sk
from bs4 import BeautifulSoup
from urllib.request import urlopen
import matplotlib.pyplot as plt
import seaborn as sns
import re
from sklearn.linear_model import LinearRegression
from sklearn.model_selection import train_test_split
from sklearn.metrics import r2_score
from sklearn.metrics import mean_squared_error
from urllib.request import urlopen
from bs4 import BeautifulSoup
import requests
import pprint
import time as tm #unit default is milliseconds from 1/1970
import datetime as dt
Display
Setting the display options can be benificial for custom viewing of data.
Col and Row Display:
#Max rows and cols; max width and height; removes the ellipsis (...) if the data is large;
#'None' here displays all data;
pd.options.display.max_columns = None
pd.options.display.max_rows = None
pd.set_option('max_colwidth', None)
pd.set_option('max_rows', None)
#Display the whole df without a new line separator (\):
pd.set_option('display.expand_frame_repr', False)
#Turn off all warnings and caveats:
warnings.filterwarnings("ignore")
#Turn off all warnings and caveats:
warnings.filterwarnings("ignore")
Data frame information:
Display basic dataframe information to give you an idea of what you are looking at. It is also extremely useful for identifying the data types of the columns—these are very important when trying to work with that data.
Basic dataframe information methods: Here is a dataframe we are to use in the following examples.
In[]: df_0 = pd.read_csv(r'path\to\file\some_file_name.csv')
In[]: df_0
Out[]:
col_1 col_2 col_3 col_4 col_5 col_6 col_7 col_8 col_9 col_10
0 Italy 332 3.38 16 461 969 260 30.0 -12 True
1 Greece 198 7.00 42 770 107 229 85.0 -65 False
2 Spain 676 0.84 45 650 82 354 51.0 -56 False
3 Rome 608 2.00 76 408 84 538 98.0 -93 True
4 South Africa 923 5.00 40 686 346 352 31.0 -37 True
5 Brazil 272 6.07 90 891 487 348 NaN 94 True
6 Romania 658 5.38 60 13 695 529 45.0 25 False
7 Japan 582 5.69 1 159 807 77 9.0 99 False
8 Canada 236 7.38 39 797 633 14 65.0 72 False
9 India 6 3.92 65 719 887 612 12.0 40 True
Get an array (list) of the columns:
'''
Call the columns into an array (list). In some instances you may run into data that
is structured with a large number of columns. Calling the columns can help pull the
data needed for whatever analysis.
'''
In[]: df_0.columns
Out[]:
Index(['col_1', 'col_2', 'col_3', 'col_4', 'col_5', 'col_6', 'col_7', 'col_8',
'col_9', 'col_10'],
dtype='object')
Call the head of the dataframe:
'''
Calling the first five rows and columns of a dataframe can be useful for an initial
look into the data. If you have a dataframe with a massive amount of rows and/or columns
it is impractal to view the entire data. Here we use a built in function that calls the
first five rows and columns.
'''
In[]: df_0.head()
Out[]:
col_1 col_2 col_3 col_4 col_5 col_6 col_7 col_8 col_9 col_10
0 Italy 332 3.38 16 461 969 260 30.0 -12 True
1 Greece 198 7.00 42 770 107 229 85.0 -65 False
2 Spain 676 0.84 45 650 82 354 51.0 -56 False
3 Rome 608 2.00 76 408 84 538 98.0 -93 True
4 South Africa 923 5.00 40 686 346 352 31.0 -37 True
Call the tail of the dataframe:
'''
Now we can use another built in function to look at the last five rows and columns.
'''
In[]: df_0.tail()
Out[]:
col_1 col_2 col_3 col_4 col_5 col_6 col_7 col_8 col_9 col_10
5 Brazil 272 6.07 90 891 487 348 NaN 94 True
6 Romania 658 5.38 60 13 695 529 45.0 25 False
7 Japan 582 5.69 1 159 807 77 9.0 99 False
8 Canada 236 7.38 39 797 633 14 65.0 72 False
9 India 6 3.92 65 719 887 612 12.0 40 True
Call the shape of the dataframe:
'''
Show the shape of the dataframe with shape.
This returns a tuple of the shape of the data.
'''
In[]: df_0.shape
Out[]: (10, 10)
Call the datatypes of each column:
'''
Now let's look at the datatypes in the dataframe with dtypes. Note that
the data types of each column may not be what it initially appears
when looking at the actual dataframe—specifically col_7 here. In the
dataframe it seems to be integers but in the actual data it is coded as
an object. This means that the data in col_7 are actually strings because
object in this return is equal to a string type data.
'''
In[]: df_0.dtypes
Out[]:
col_1 object
col_2 int64
col_3 float64
col_4 int64
col_5 int64
col_6 int64
col_7 object
col_8 float64
col_9 int64
col_10 bool
dtype: object
Call detailed information of each column:
'''
Get more detailed information on the data with info(). When looking
at the return of info() we see that the columns also have an index (left).
Next, follows the column names. Followed by the non-null counts of each
column—meaning our data frame is shaped (10,10) so of all the data col_8
has 1 null data cell. Null (NaN) data represents missing data.
'''
In[]: df_0.info()
Out[]:
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 10 entries, 0 to 9
Data columns (total 10 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 col_1 10 non-null object
1 col_2 10 non-null int64
2 col_3 10 non-null float64
3 col_4 10 non-null int64
4 col_5 10 non-null int64
5 col_6 10 non-null int64
6 col_7 10 non-null object
7 col_8 9 non-null float64
8 col_9 10 non-null int64
9 col_10 10 non-null bool
dtypes: bool(1), float64(2), int64(5), object(2)
memory usage: 858.0+ bytes
Basic Dataframe Index Slicing With .loc and .iloc
Index slicing will be very useful for scoping in on particular data.
If we look at how a dataframe is structured by its index we can see what
are essentially grid coordinates of each cell, row, and column.
Accessing data in a dataframe allows one to apply methods for xyz analysis to that data.
For example adding one column to another column to generate a third column, applying
an operator for statistical analysis, or converting a column based on some condition. These
are only a few examples of many.
Here I will start very simple with df_0 (same dataframe as above).
So, here again is our dataframe for reference:
In[]: df_0
Out[]:
col_1 col_2 col_3 col_4 col_5 col_6 col_7 col_8 col_9 col_10
0 Italy 332 3.38 16 461 969 260 30.0 -12 True
1 Greece 198 7.00 42 770 107 229 85.0 -65 False
2 Spain 676 0.84 45 650 82 354 51.0 -56 False
3 Rome 608 2.00 76 408 84 538 98.0 -93 True
4 South Africa 923 5.00 40 686 346 352 31.0 -37 True
5 Brazil 272 6.07 90 891 487 348 NaN 94 True
6 Romania 658 5.38 60 13 695 529 45.0 25 False
7 Japan 582 5.69 1 159 807 77 9.0 99 False
8 Canada 236 7.38 39 797 633 14 65.0 72 False
9 India 6 3.92 65 719 887 612 12.0 40 True
Accessing Specific Cells Based on Index:
Access a single cell:
'''
The basic syntax for .iloc is SomeDataframe.iloc[Row_Index, Col_Index].
'''
In[]: df_0.iloc[0,0]
Out[]: 'Italy'
In[]: df_0.iloc[9,0]
Out[]: 'India'
In[]: df_0.iloc[0,4]
Out[]: 461
#remember 'col_7' at, axis 1 index 6, are string objects not integers;
#I will show methods to convert data types later
In[]: df_0.iloc[0,6]
Out[]: '260'
'''
You can now assign specific data to other variable objects too.
'''
In[]: data_var_1 = df_0.iloc[0,4]
In[]: data_var_1 * 2 / 3
Out[]: 307.3333333333333
Access a group of cells:
#you can think of the syntax like; SomeDataframe[Row_idx_start:Row_idx_stop, Col_idx_start:Col_idx_stop]
In[]: df_0.iloc[0:2,0:4]
Out[]:
col_1 col_2 col_3 col_4
0 Italy 332 3.38 16
1 Greece 198 7.00 42
In[]: df_0.iloc[3:6,3:5]
Out[]:
col_4 col_5
3 76 408
4 40 686
5 90 891
Accessing Specific Rows Based on Index:
Accessing multipul rows:
#access the first three rows
In[]: df_0.iloc[:2]
Out[]:
col_1 col_2 col_3 col_4 col_5 col_6 col_7 col_8 col_9 col_10
0 Italy 332 3.38 16 461 969 260 30.0 -12 True
1 Greece 198 7.00 42 770 107 229 85.0 -65 False
2 Spain 676 0.84 45 650 82 354 51.0 -56 False
#access all rows after nth row
In[]: df_0.iloc[7:]
Out[]:
col_1 col_2 col_3 col_4 col_5 col_6 col_7 col_8 col_9 col_10
7 Japan 582 5.69 1 159 807 77 9.0 99 False
8 Canada 236 7.38 39 797 633 14 65.0 72 False
9 India 6 3.92 65 719 887 612 12.0 40 True
#access rows at row index four, five, and six
In[]: df_0.iloc[4:7]
Out[]:
col_1 col_2 col_3 col_4 col_5 col_6 col_7 col_8 col_9 col_10
4 South Africa 923 5.00 40 686 346 352 31.0 -37 True
5 Brazil 272 6.07 90 891 487 348 NaN 94 True
6 Romania 658 5.38 60 13 695 529 45.0 25 False
#access all rows except the last nth number
In[]: df_0.iloc[:-4]
Out[]:
col_1 col_2 col_3 col_4 col_5 col_6 col_7 col_8 col_9 col_10
0 Italy 332 3.38 16 461 969 260 30.0 -12 True
1 Greece 198 7.00 42 770 107 229 85.0 -65 False
2 Spain 676 0.84 45 650 82 354 51.0 -56 False
3 Rome 608 2.00 76 408 84 538 98.0 -93 True
4 South Africa 923 5.00 40 686 346 352 31.0 -37 True
5 Brazil 272 6.07 90 891 487 348 NaN 94 True
#access every second index rows starting at 0
In[]: df_0.iloc[::2]
Out[]:
col_1 col_2 col_3 col_4 col_5 col_6 col_7 col_8 col_9 col_10
0 Italy 332 3.38 16 461 969 260 30.0 -12 True
2 Spain 676 0.84 45 650 82 354 51.0 -56 False
4 South Africa 923 5.00 40 686 346 352 31.0 -37 True
6 Romania 658 5.38 60 13 695 529 45.0 25 False
8 Canada 236 7.38 39 797 633 14 65.0 72 False
#access every third index rows starting at 0
In[]: df_0.iloc[::3]
Out[]:
col_1 col_2 col_3 col_4 col_5 col_6 col_7 col_8 col_9 col_10
0 Italy 332 3.38 16 461 969 260 30.0 -12 True
3 Rome 608 2.00 76 408 84 538 98.0 -93 True
6 Romania 658 5.38 60 13 695 529 45.0 25 False
9 India 6 3.92 65 719 887 612 12.0 40 True
#access all nth rows beggining at the end of row index
In[]: df_0.iloc[::-2]
Out[]:
col_1 col_2 col_3 col_4 col_5 col_6 col_7 col_8 col_9 col_10
9 India 6 3.92 65 719 887 612 12.0 40 True
7 Japan 582 5.69 1 159 807 77 9.0 99 False
5 Brazil 272 6.07 90 891 487 348 NaN 94 True
3 Rome 608 2.00 76 408 84 538 98.0 -93 True
1 Greece 198 7.00 42 770 107 229 85.0 -65 False
'''
Assign an index slice to a variable object.
'''
In[]: evens_df_0 = df_0.iloc[::2]
In[]: evens_df_0
Out[]:
col_1 col_2 col_3 col_4 col_5 col_6 col_7 col_8 col_9 col_10
0 Italy 332 3.38 16 461 969 260 30.0 -12 True
2 Spain 676 0.84 45 650 82 354 51.0 -56 False
4 South Africa 923 5.00 40 686 346 352 31.0 -37 True
6 Romania 658 5.38 60 13 695 529 45.0 25 False
8 Canada 236 7.38 39 797 633 14 65.0 72 False
Accessing Specific Columns Based on Index:
#access a single column as a series
In[]: df_0.iloc[0:,0]
Out[]:
0 Italy
1 Greece
2 Spain
3 Rome
4 South Africa
5 Brazil
6 Romania
7 Japan
8 Canada
9 India
Name: col_1, dtype: object
#accessing a single column as a dataframe
In[]: df_0.iloc[0:,0:1]
Out[]:
col_1
0 Italy
1 Greece
2 Spain
3 Rome
4 South Africa
5 Brazil
6 Romania
7 Japan
8 Canada
9 India
#access several columns
In[]: df_0.iloc[0:,2:5]
Out[]:
col_3 col_4 col_5
0 3.38 16 461
1 7.00 42 770
2 0.84 45 650
3 2.00 76 408
4 5.00 40 686
5 6.07 90 891
6 5.38 60 13
7 5.69 1 159
8 7.38 39 797
9 3.92 65 719
#access all columns after specified index
In[]: df_0.iloc[0:,7:]
Out[]:
col_8 col_9 col_10
0 30.0 -12 True
1 85.0 -65 False
2 51.0 -56 False
3 98.0 -93 True
4 31.0 -37 True
5 NaN 94 True
6 45.0 25 False
7 9.0 99 False
8 65.0 72 False
9 12.0 40 True
#access every nth columns
In[]: df_0.iloc[0:,0::2]
Out[]:
col_1 col_3 col_5 col_7 col_9
0 Italy 3.38 461 260 -12
1 Greece 7.00 770 229 -65
2 Spain 0.84 650 354 -56
3 Rome 2.00 408 538 -93
4 South Africa 5.00 686 352 -37
5 Brazil 6.07 891 348 94
6 Romania 5.38 13 529 25
7 Japan 5.69 159 77 99
8 Canada 7.38 797 14 72
9 India 3.92 719 612 40
Other Slicing Methods
Here I include some methods—outside of .loc and iloc—that I found very useful for isolating rows or cols.
Again, here is the example dataframe we will be using:
In[]: df_0
Out[]:
col_1 col_2 col_3 col_4 col_5 col_6 col_7 col_8 col_9 col_10
0 Italy 332 3.38 16 461 969 260 30.0 -12 True
1 Greece 198 7.00 42 770 107 229 85.0 -65 False
2 Spain 676 0.84 45 650 82 354 51.0 -56 False
3 Rome 608 2.00 76 408 84 538 98.0 -93 True
4 South Africa 923 5.00 40 686 346 352 31.0 -37 True
5 Brazil 272 6.07 90 891 487 348 NaN 94 True
6 Romania 658 5.38 60 13 695 529 45.0 25 False
7 Japan 582 5.69 1 159 807 77 9.0 99 False
8 Canada 236 7.38 39 797 633 14 65.0 72 False
9 India 6 3.92 65 719 887 612 12.0 40 True
Slicing a dataframe by column names:
#use the column names
In[]: new_df = df_0[['col_1', 'col_3', 'col_9']]
In[]: new_df
Out[]:
col_1 col_3 col_9
0 Italy 3.38 -12
1 Greece 7.00 -65
2 Spain 0.84 -56
3 Rome 2.00 -93
4 South Africa 5.00 -37
5 Brazil 6.07 94
6 Romania 5.38 25
7 Japan 5.69 99
8 Canada 7.38 72
9 India 3.92 40
Slice rows by row index:
'''
This does not change the data unless you re-assign to
a new object variable or add the argument inplace=True
'''
In[]: df_0.drop([0,1,2,3]) #df_0.drop([0,1,2,3], inplace=True) ; makes a permanent change
Out[]:
col_1 col_2 col_3 col_4 col_5 col_6 col_7 col_8 col_9 col_10
4 South Africa 923 5.00 40 686 346 352 31.0 -37 True
5 Brazil 272 6.07 90 891 487 348 NaN 94 True
6 Romania 658 5.38 60 13 695 529 45.0 25 False
7 Japan 582 5.69 1 159 807 77 9.0 99 False
8 Canada 236 7.38 39 797 633 14 65.0 72 False
9 India 6 3.92 65 719 887 612 12.0 40 True
Use a list as an index filter:
'''
You generate you own list of integers to filter the index as well. Here, this
drops all the even indexed rows.
'''
In[]: idx_every_two = [i for i in range(0,9,2)]
In[]: df_0.drop(idx_every_two)
Out[]:
col_1 col_2 col_3 col_4 col_5 col_6 col_7 col_8 col_9 col_10
1 Greece 198 7.00 42 770 107 229 85.0 -65 False
3 Rome 608 2.00 76 408 84 538 98.0 -93 True
5 Brazil 272 6.07 90 891 487 348 NaN 94 True
7 Japan 582 5.69 1 159 807 77 9.0 99 False
9 India 6 3.92 65 719 887 612 12.0 40 True
Conditional Index Slicing
Conditional index slicing can get rather complex because some of the syntax is not very intuitive.
Here again is our dataframe:
In[]: df_0
Out[]:
col_1 col_2 col_3 col_4 col_5 col_6 col_7 col_8 col_9 col_10
0 Italy 332 3.38 16.0 461 969.0 260 30.0 -12 True
1 Greece 198 7.00 42.0 770 107.0 229 85.0 -65 False
2 Spain 676 0.84 45.0 650 82.0 354 51.0 -56 False
3 Rome 608 2.00 76.0 408 84.0 538 98.0 -93 True
4 South Africa 923 5.00 40.0 686 346.0 352 31.0 -37 True
6 Romania 658 5.38 60.0 13 695.0 529 45.0 25 False
7 Japan 582 5.69 1.0 159 807.0 77 9.0 99 False
8 Canada 236 7.38 39.0 797 633.0 14 65.0 72 False
9 India 6 3.92 65.0 719 887.0 612 12.0 40 True
Slice rows based on one columns values:
'''
Here we select all rows where col_5 is less than 500. If the data in the column is
numeric you can implement any comparison operator to perform a conditional extraction
of data. If it is string data you can still perform operations that are valid for that data.
'''
In[]: df_0.loc[df_0['col_5'] < 500]
Out[]:
col_1 col_2 col_3 col_4 col_5 col_6 col_7 col_8 col_9 col_10
0 Italy 332 3.38 16 461 969 260 30.0 -12 True
3 Rome 608 2.00 76 408 84 538 98.0 -93 True
6 Romania 658 5.38 60 13 695 529 45.0 25 False
7 Japan 582 5.69 1 159 807 77 9.0 99 False
'''
Here is another example useing col_10 by selecting all rows where col_10 is True
'''
In[]: df_0.loc[df_0['col_10'] == True]
Out[]:
col_1 col_2 col_3 col_4 col_5 col_6 col_7 col_8 col_9 col_10
0 Italy 332 3.38 16 461 969 260 30.0 -12 True
3 Rome 608 2.00 76 408 84 538 98.0 -93 True
4 South Africa 923 5.00 40 686 346 352 31.0 -37 True
5 Brazil 272 6.07 90 891 487 348 NaN 94 True
9 India 6 3.92 65 719 887 612 12.0 40 True
Slice rows based on multipul columns and data conditions:
'''
Here we will slice data where it is greater than 500 in col_2 and False in col_10
'''
In[]: df_0.loc[(df_0['col_2'] > 500) & (df_0['col_10'] == False)]
Out[]:
col_1 col_2 col_3 col_4 col_5 col_6 col_7 col_8 col_9 col_10
2 Spain 676 0.84 45 650 82 354 51.0 -56 False
6 Romania 658 5.38 60 13 695 529 45.0 25 False
7 Japan 582 5.69 1 159 807 77 9.0 99 False
Slice a dataframe based on string column values:
'''
Here I used the string method startswith() but you can use some other string methods.
'''
In[]: df_0.loc[df_0['col_1'].str.startswith("I")]
Out[]:
col_1 col_2 col_3 col_4 col_5 col_6 col_7 col_8 col_9 col_10
0 Italy 332 3.38 16 461 969 260 30.0 -12 True
9 India 6 3.92 65 719 887 612 12.0 40 True
Slice a dataframe based a numeric condition and a alphabetical condition:
'''
A string that starts with 'I' in col_1 and a integer value less that 50 in col_4.
'''
In[]: df_0.loc[(df_0['col_1'].str.startswith("I")) & (df_0['col_4'] < 50)]
Out[]:
col_1 col_2 col_3 col_4 col_5 col_6 col_7 col_8 col_9 col_10
0 Italy 332 3.38 16 461 969 260 30.0 -12 True
'''
Returns rows that endwith 'a' in col_1 and are
less than 100 in col_4 and less than 80 in col_9.
'''
In[]: df_0.loc[(df_0['col_1'].str.endswith("a")) & (df_0['col_4'] < 100) & (df_0['col_9'] < 80)]
Out[]:
col_1 col_2 col_3 col_4 col_5 col_6 col_7 col_8 col_9 col_10
4 South Africa 923 5.00 40 686 346 352 31.0 -37 True
6 Romania 658 5.38 60 13 695 529 45.0 25 False
8 Canada 236 7.38 39 797 633 14 65.0 72 False
9 India 6 3.92 65 719 887 612 12.0 40 True
Applying Operators to Columns And Rows
Applying operations to columns and rows may be needed for a many different reasons. Once you know how to access the data applying operations to it becomes very easy.
Here again is our dataframe:
In[]: df_0
Out[]:
col_1 col_2 col_3 col_4 col_5 col_6 col_7 col_8 col_9 col_10
0 Italy 332 3.38 16.0 461 969.0 260 30.0 -12 True
1 Greece 198 7.00 42.0 770 107.0 229 85.0 -65 False
2 Spain 676 0.84 45.0 650 82.0 354 51.0 -56 False
3 Rome 608 2.00 76.0 408 84.0 538 98.0 -93 True
4 South Africa 923 5.00 40.0 686 346.0 352 31.0 -37 True
6 Romania 658 5.38 60.0 13 695.0 529 45.0 25 False
7 Japan 582 5.69 1.0 159 807.0 77 9.0 99 False
8 Canada 236 7.38 39.0 797 633.0 14 65.0 72 False
9 India 6 3.92 65.0 719 887.0 612 12.0 40 True
Apply operators to one column:
'''
Add 100 to col_4
'''
In[]: df_0['col_4'] += 100
In[]: df_0
Out[]:
col_1 col_2 col_3 col_4 col_5 col_6 col_7 col_8 col_9 col_10
0 Italy 332 3.38 116 461 969 260 30.0 -12 True
1 Greece 198 7.00 142 770 107 229 85.0 -65 False
2 Spain 676 0.84 145 650 82 354 51.0 -56 False
3 Rome 608 2.00 176 408 84 538 98.0 -93 True
4 South Africa 923 5.00 140 686 346 352 31.0 -37 True
5 Brazil 272 6.07 190 891 487 348 NaN 94 True
6 Romania 658 5.38 160 13 695 529 45.0 25 False
7 Japan 582 5.69 101 159 807 77 9.0 99 False
8 Canada 236 7.38 139 797 633 14 65.0 72 False
9 India 6 3.92 165 719 887 612 12.0 40 True
'''
Divide a column.
'''
In[]: df_0['col_3'] = df_0['col_3'] / 100
In[]: df_0
Out[]:
col_1 col_2 col_3 col_4 col_5 col_6 col_7 col_8 col_9 col_10
0 Italy 332 0.0338 136.0 461 969 260 30.0 -12 True
1 Greece 198 0.0700 162.0 770 107 229 85.0 -65 False
2 Spain 676 0.0084 165.0 650 82 354 51.0 -56 False
3 Rome 608 0.0200 196.0 408 84 538 98.0 -93 True
4 South Africa 923 0.0500 160.0 686 346 352 31.0 -37 True
5 Brazil 272 0.0607 210.0 891 487 348 NaN 94 True
6 Romania 658 0.0538 180.0 13 695 529 45.0 25 False
7 Japan 582 0.0569 121.0 159 807 77 9.0 99 False
8 Canada 236 0.0738 159.0 797 633 14 65.0 72 False
9 India 6 0.0392 185.0 719 887 612 12.0 40 True
Access data with .iloc and apply an operator:
'''
From the original data;
Here we added 100 to each data point in columns [col_2, col_3, col_4, col_5]
'''
In[]: df_0.iloc[0:,1:5] += 100
In[]: df_0
Out[]:
col_1 col_2 col_3 col_4 col_5 col_6 col_7 col_8 col_9 col_10
0 Italy 432 103.38 116 561 969 260 30.0 -12 True
1 Greece 298 107.00 142 870 107 229 85.0 -65 False
2 Spain 776 100.84 145 750 82 354 51.0 -56 False
3 Rome 708 102.00 176 508 84 538 98.0 -93 True
4 South Africa 1023 105.00 140 786 346 352 31.0 -37 True
5 Brazil 372 106.07 190 991 487 348 NaN 94 True
6 Romania 758 105.38 160 113 695 529 45.0 25 False
7 Japan 682 105.69 101 259 807 77 9.0 99 False
8 Canada 336 107.38 139 897 633 14 65.0 72 False
9 India 106 103.92 165 819 887 612 12.0 40 True
Create Columns Based on Conditions Of Other Columns
Here are examples of things you may need to do to anayze data based on conditions. Most of the time each column represents a variable of some kind. These variables can be used to craft other calculations and variables.
Here again is our dataframe:
In[]: df_0
Out[]:
col_1 col_2 col_3 col_4 col_5 col_6 col_7 col_8 col_9 col_10
0 Italy 332 3.38 16 461 969 260 30.0 -12 True
1 Greece 198 7.00 42 770 107 229 85.0 -65 False
2 Spain 676 0.84 45 650 82 354 51.0 -56 False
3 Rome 608 2.00 76 408 84 538 98.0 -93 True
4 South Africa 923 5.00 40 686 346 352 31.0 -37 True
5 Brazil 272 6.07 90 891 487 348 NaN 94 True
6 Romania 658 5.38 60 13 695 529 45.0 25 False
7 Japan 582 5.69 1 159 807 77 9.0 99 False
8 Canada 236 7.38 39 797 633 14 65.0 72 False
9 India 6 3.92 65 719 887 612 12.0 40 True
'''
Say we need to find all the values in col_5 that are greater than values in col_2.
Here is how we create a new column (col_11) to display the results across each
columns respective rows as True if col_5 is greater than col_2 and False if col_2
is less than col_5.
'''
In[]: df_0['col_11'] = df_0['col_5'] > df_0['col_2']
In[]: df_0
Out[]:
col_1 col_2 col_3 col_4 col_5 col_6 col_7 col_8 col_9 col_10 col_11
0 Italy 432 103.38 116 561 969 260 30.0 -12 True True
1 Greece 298 107.00 142 870 107 229 85.0 -65 False True
2 Spain 776 100.84 145 750 82 354 51.0 -56 False False
3 Rome 708 102.00 176 508 84 538 98.0 -93 True False
4 South Africa 1023 105.00 140 786 346 352 31.0 -37 True False
5 Brazil 372 106.07 190 991 487 348 NaN 94 True True
6 Romania 758 105.38 160 113 695 529 45.0 25 False False
7 Japan 682 105.69 101 259 807 77 9.0 99 False False
8 Canada 336 107.38 139 897 633 14 65.0 72 False True
9 India 106 103.92 165 819 887 612 12.0 40 True True
'''
Or, say we need to add a column that is the sum of col_3 and col_4 divided by 100.
'''
In[]: df_0['col_12'] = (df_0['col_3'] + df_0['col_4']) / 100
In[]: df_0
Out[]:
col_1 col_2 col_3 col_4 col_5 col_6 col_7 col_8 col_9 col_10 col_11 col_12
0 Italy 332 3.38 16 461 969 260 30.0 -12 True True 0.1938
1 Greece 198 7.00 42 770 107 229 85.0 -65 False True 0.4900
2 Spain 676 0.84 45 650 82 354 51.0 -56 False False 0.4584
3 Rome 608 2.00 76 408 84 538 98.0 -93 True False 0.7800
4 South Africa 923 5.00 40 686 346 352 31.0 -37 True False 0.4500
5 Brazil 272 6.07 90 891 487 348 NaN 94 True True 0.9607
6 Romania 658 5.38 60 13 695 529 45.0 25 False False 0.6538
7 Japan 582 5.69 1 159 807 77 9.0 99 False False 0.0669
8 Canada 236 7.38 39 797 633 14 65.0 72 False True 0.4638
9 India 6 3.92 65 719 887 612 12.0 40 True True 0.6892
Merging Dataframes
Merging dataframes is also known as concatination.
'''
Here are two data frames that we can merge—df_3 and df_4.
'''
In[]: df_3
Out[]:
datetime col_1 col_2
0 2009-1-20 1 22
1 2009-2-23 2 23
2 2009-8-24 3 24
3 2010-7-24 4 25
4 2010-8-22 5 26
5 2010-7-21 6 27
6 2010-8-15 7 28
7 2011-4-22 8 29
8 2011-2-22 9 30
9 2011-8-25 10 31
In[]: df_4
Out[]:
country date var_1 var_2
0 Italy 2009-1-20 3.38 16
1 Greece 2009-2-23 7.00 42
2 Spain 2009-8-24 1.90 66
3 Rome 2010-7-24 2.00 76
4 South Africa 2010-8-22 5.00 40
5 Brazil 2010-7-21 6.07 22
6 Romania 2010-8-15 5.38 60
7 Japan 2011-4-22 5.69 1
8 Canada 2011-2-22 7.38 39
9 India 2011-8-25 3.92 65
'''
One easy way to merge df_3 and df_4 is by index.
'''
In[]: df_4.merge(df_3, left_index=True, right_index=True, how='right')
Out[]:
country date var_1 var_2 datetime col_1 col_2
0 Italy 2009-1-20 3.38 16 2009-1-20 1 22
1 Greece 2009-2-23 7.00 42 2009-2-23 2 23
2 Spain 2009-8-24 1.90 66 2009-8-24 3 24
3 Rome 2010-7-24 2.00 76 2010-7-24 4 25
4 South Africa 2010-8-22 5.00 40 2010-8-22 5 26
5 Brazil 2010-7-21 6.07 22 2010-7-21 6 27
6 Romania 2010-8-15 5.38 60 2010-8-15 7 28
7 Japan 2011-4-22 5.69 1 2011-4-22 8 29
8 Canada 2011-2-22 7.38 39 2011-2-22 9 30
9 India 2011-8-25 3.92 65 2011-8-25 10 31
'''
You might run into the dates being out of order on one of the dataframes.
But the column that is being used to merge must have the same name in both
dataframes.
Like this:
'''
In[]: df_3
Out[]:
datetime col_1 col_2
0 2010-8-22 1 22
1 2009-2-23 2 23
2 2011-8-25 3 24
3 2011-2-22 4 25
4 2009-8-24 5 26
5 2009-1-20 6 27
6 2010-7-24 7 28
7 2010-7-21 8 29
8 2010-8-15 9 30
9 2011-4-22 10 31
In[]: df_4
Out[]:
country datetime var_1 var_2
0 Italy 2009-1-20 3.38 16
1 Greece 2009-2-23 7.00 42
2 Spain 2009-8-24 1.90 66
3 Rome 2010-7-24 2.00 76
4 South Africa 2010-8-22 5.00 40
5 Brazil 2010-7-21 6.07 22
6 Romania 2010-8-15 5.38 60
7 Japan 2011-4-22 5.69 1
8 Canada 2011-2-22 7.38 39
9 India 2011-8-25 3.92 65
#Merge by datetime column.
In[]: df_4.merge(df_3, on='datetime', how='right')
Out[]:
country datetime var_1 var_2 col_1 col_2
0 South Africa 2010-8-22 5.00 40 1 22
1 Greece 2009-2-23 7.00 42 2 23
2 India 2011-8-25 3.92 65 3 24
3 Canada 2011-2-22 7.38 39 4 25
4 Spain 2009-8-24 1.90 66 5 26
5 Italy 2009-1-20 3.38 16 6 27
6 Rome 2010-7-24 2.00 76 7 28
7 Brazil 2010-7-21 6.07 22 8 29
8 Romania 2010-8-15 5.38 60 9 30
9 Japan 2011-4-22 5.69 1 10 31
Data Cleaning
You often will not start with a clean dataframe. Meaning that you will
be required to do some cleaning of the data—that is without impacting its
validitiy. One of the major goals here is re-craft the data's uniformity.
You want to be working with uniform data. So, for these examples I will use
a new dataframe that is dirty and clean it up.
You often will not start with clean data—especially if the data was scraped from
a website.
Here is a dirty dataframe example:
In[]: dirty_df
Out[]:
col_1 col_2 col_3 col_4 col_5 col_6 col_7 col_8 col_9
0 Italy()2 332 3.38 16.0 461 969.0 260 30.0 -12
1 GrEECE(1) 198 7.00 42.0 770 107.0 229 85.0 -65
2 (Spain) 676 NaN NaN 650 82.0 354 51.0 -56
3 !.Rom e -- 2.00 76.0 408 84.0 538 98.0 -93
4 sOuth A!!frica 923 5.00 40.0 686 346.0 352 31.0 -37
5 (11)Brazil 272 6.07 NaN 891 NaN 348 NaN 94
6 *Romania 658 5.38 60.0 13 695.0 529 45.0 25
7 Japan00 582 5.69 1.0 159 807.0 77 9.0 99
8 Canada6 236 7.38 39.0 797 633.0 14 65.0 72
9 !!India 6 3.92 65.0 719 887.0 612 12.0 40
'''
Let's start with looking at the info() of the dataframe.
'''
In[]: dirty_df.info()
Out[]:
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 10 entries, 0 to 9
Data columns (total 10 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 col_1 10 non-null object
1 col_2 10 non-null object #because there is one string data in col_2 it shows as object data
2 col_3 9 non-null float64
3 col_4 8 non-null float64
4 col_5 10 non-null int64
5 col_6 9 non-null float64
6 col_7 10 non-null object
7 col_8 9 non-null float64
8 col_9 10 non-null int64
dtypes: bool(1), float64(4), int64(2), object(3)
memory usage: 858.0+ bytes
'''
Let's start with col_1 and strip all the unwanted string vals and attain only the countries strings.
'''
#first lets remove the digits from the data
#the following line uses regex to replace anything that isn't a letter or integer
In[]: dirty_df['col_1'].replace('\W+', '',regex=True,inplace=True)
In[]: dirty_df
Out[]:
col_1 col_2 col_3 col_4 col_5 col_6 col_7 col_8 col_9
0 Italy2 332 3.38 16.0 461 969.0 260 30.0 -12
1 GrEECE1 198 7.00 42.0 770 107.0 229 85.0 -65
2 Spain 676 NaN NaN 650 82.0 354 51.0 -56
3 Rome -- 2.00 76.0 408 84.0 538 98.0 -93
4 sOuthAfrica 923 5.00 40.0 686 346.0 352 31.0 -37
5 11Brazil 272 6.07 NaN 891 NaN 348 NaN 94
6 Romania 658 5.38 60.0 13 695.0 529 45.0 25
7 Japan00 582 5.69 1.0 159 807.0 77 9.0 99
8 Canada6 236 7.38 39.0 797 633.0 14 65.0 72
9 India 6 3.92 65.0 719 887.0 612 12.0 40
#now remove all the unwanted trailing or leading numbers with the same method
In[]: dirty_df['col_1'].replace('\d+', '',regex=True,inplace=True)
In[]: dirty_df
Out[]:
col_1 col_2 col_3 col_4 col_5 col_6 col_7 col_8 col_9
0 Italy 332 3.38 16.0 461 969.0 260 30.0 -12
1 GrEECE 198 7.00 42.0 770 107.0 229 85.0 -65
2 Spain 676 NaN NaN 650 82.0 354 51.0 -56
3 Rome -- 2.00 76.0 408 84.0 538 98.0 -93
4 sOuthAfrica 923 5.00 40.0 686 346.0 352 31.0 -37
5 Brazil 272 6.07 NaN 891 NaN 348 NaN 94
6 Romania 658 5.38 60.0 13 695.0 529 45.0 25
7 Japan 582 5.69 1.0 159 807.0 77 9.0 99
8 Canada 236 7.38 39.0 797 633.0 14 65.0 72
9 India 6 3.92 65.0 719 887.0 612 12.0 40
'''
Now let's make col_1's case uniform. Here we use upper().
'''
In[]: dirty_df['col_1'] = dirty_df['col_1'].str.upper()
In[]: dirty_df
Out[]:
col_1 col_2 col_3 col_4 col_5 col_6 col_7 col_8 col_9
0 ITALY 332 3.38 16.0 461 969.0 260 30.0 -12
1 GREECE 198 7.00 42.0 770 107.0 229 85.0 -65
2 SPAIN 676 NaN NaN 650 82.0 354 51.0 -56
3 ROME -- 2.00 76.0 408 84.0 538 98.0 -93
4 SOUTHAFRICA 923 5.00 40.0 686 346.0 352 31.0 -37
5 BRAZIL 272 6.07 NaN 891 NaN 348 NaN 94
6 ROMANIA 658 5.38 60.0 13 695.0 529 45.0 25
7 JAPAN 582 5.69 1.0 159 807.0 77 9.0 99
8 CANADA 236 7.38 39.0 797 633.0 14 65.0 72
9 INDIA 6 3.92 65.0 719 887.0 612 12.0 40
'''
Now convert the NaN data. This method will change all the Nan values to 0.0.
You may need to do this during an analysis because the dataframe needs to be
equally shaped—meaning, in this instance, you can't have less than 10 items in
a column. Changing the Nan values to 0 usually won't invalidate the data and
allows you to perform other operations—because it doesn't change the shape of
the distribution.
'''
In[]: dirty_df.fillna(0, inplace=True)
In[]: dirty_df
Out[]:
col_1 col_2 col_3 col_4 col_5 col_6 col_7 col_8 col_9
0 ITALY 332 3.38 16.0 461 969.0 260 30.0 -12
1 GREECE 198 7.00 42.0 770 107.0 229 85.0 -65
2 SPAIN 676 0.00 0.0 650 82.0 354 51.0 -56
3 ROME -- 2.00 76.0 408 84.0 538 98.0 -93
4 SOUTHAFRICA 923 5.00 40.0 686 346.0 352 31.0 -37
5 BRAZIL 272 6.07 0.0 891 0.0 348 0.0 94
6 ROMANIA 658 5.38 60.0 13 695.0 529 45.0 25
7 JAPAN 582 5.69 1.0 159 807.0 77 9.0 99
8 CANADA 236 7.38 39.0 797 633.0 14 65.0 72
9 INDIA 6 3.92 65.0 719 887.0 612 12.0 40
'''
Finally let's get replace the '--' in col_2 (which could be unreported data) with 0.
'''
In[]: dirty_df['col_2'].replace('--', '0',regex=True,inplace=True)
In[]: dirty_df
Out[]:
col_1 col_2 col_3 col_4 col_5 col_6 col_7 col_8 col_9
0 ITALY 332 3.38 16.0 461 969.0 260 30.0 -12
1 GREECE 198 7.00 42.0 770 107.0 229 85.0 -65
2 SPAIN 676 0.00 0.0 650 82.0 354 51.0 -56
3 ROME 0 2.00 76.0 408 84.0 538 98.0 -93
4 SOUTHAFRICA 923 5.00 40.0 686 346.0 352 31.0 -37
5 BRAZIL 272 6.07 0.0 891 0.0 348 0.0 94
6 ROMANIA 658 5.38 60.0 13 695.0 529 45.0 25
7 JAPAN 582 5.69 1.0 159 807.0 77 9.0 99
8 CANADA 236 7.38 39.0 797 633.0 14 65.0 72
9 INDIA 6 3.92 65.0 719 887.0 612 12.0 40
Converting Data:
'''
Here I'll use a new df we'll assign it to varible object df_1.
Here's the view of it.
'''
In[]: df_1
Out[]:
col_1 col_2 col_3 col_4 col_5 col_6 col_7 col_8 col_9
0 ITALY 332 3.38 16.0 461 969.0 260 30.0 -12
1 GREECE 198 7.00 42.0 770 107.0 229 85.0 -65
2 SPAIN 676 0.00 0.0 650 82.0 354 51.0 -56
3 ROME 0 2.00 76.0 408 84.0 538 98.0 -93
4 SOUTHAFRICA 923 5.00 40.0 686 346.0 352 31.0 -37
5 BRAZIL 272 6.07 0.0 891 0.0 348 0.0 94
6 ROMANIA 658 5.38 60.0 13 695.0 529 45.0 25
7 JAPAN 582 5.69 1.0 159 807.0 77 9.0 99
8 CANADA 236 7.38 39.0 797 633.0 14 65.0 72
9 INDIA 6 3.92 65.0 719 887.0 612 12.0 40
'''
Let's look at the data types of the columns.
'''
In[]: df_1.dtypes
Out[]:
col_1 object
col_2 object
col_3 float64
col_4 float64
col_5 int64
col_6 float64
col_7 object
col_8 float64
col_9 int64
dtype: object
'''
We can see that col_7 is a string object datatype. We need to convert
it to an integer or float type to use integer or float methods on the
data. There are few ways to do this.
'''
In[]: df_1.col_7 = df_1.col_7.astype(int) #dot syntax method
In[]: df_1.dtypes
Out[]:
col_1 object
col_2 object
col_3 float64
col_4 float64
col_5 int64
col_6 float64
col_7 int32 #col_7 data are now integers
col_8 float64
col_9 int64
dtype: object
'''
Here's an alternative syntax; changes the data to float data.
'''
In[]: df_1['col_7'] = df_1['col_7'].astype(float)
In[]: df_1.dtypes
Out[]:
col_1 object
col_2 object
col_3 float64
col_4 float64
col_5 int64
col_6 float64
col_7 float64 #col_7 data are now floats
col_8 float64
col_9 int64
dtype: object
Basic Date Time Methods
I have found datetime data to be a bit tricky but I have included here some methods to work with it.
Here is our date time dataframe:
'''
Our datetime column here is actually string data.
'''
In[]: time_df
Out[]:
col_1 datetime col_3 col_4 col_5 col_6 col_7 col_8
0 Italy 2009-1-20 3.38 16 461 969 260 30.0
1 Greece 2009-2-23 7.00 42 770 107 229 85.0
2 Spain 2009-8-24 1.90 66 650 82 354 51.0
3 Rome 2010-7-24 2.00 76 408 84 538 98.0
4 South Africa 2010-8-22 5.00 40 686 346 352 31.0
5 Brazil 2010-7-21 6.07 22 891 33 348 55.0
6 Romania 2010-8-15 5.38 60 13 695 529 45.0
7 Japan 2011-4-22 5.69 1 159 807 77 9.0
8 Canada 2011-2-22 7.38 39 797 633 14 65.0
9 India 2011-8-25 3.92 65 719 887 61 12.0
'''
First look at the data types of the columns.
'''
In[]: time_df.dtypes
Out[]:
col_1 object
datetime object #datetime is currently string data
col_3 float64
col_4 int64
col_5 int64
col_6 int64
col_7 int64
col_8 float64
dtype: object
Convert a column to datetime data:
'''
The datetime column is currently a string object.
Convert it to a "date time" data type.
'''
In[]: time_df.datetime = pd.to_datetime(time_df['datetime'], format='%Y-%m-%d')
In[]: time_df.dtypes
Out[]:
col_1 object
datetime datetime64[ns]
col_3 float64
col_4 int64
col_5 int64
col_6 int64
col_7 int64
col_8 float64
dtype: object
Slice the rows by specific year:
'''
Here we will slice the data by the year 2010.
'''
In[]: time_df.loc[(time_df["datetime"].dt.year == 2010)]
Out[]:
col_1 datetime col_3 col_4 col_5 col_6 col_7 col_8
3 Rome 2010-07-24 2.00 76 408 84 538 98.0
4 South Africa 2010-08-22 5.00 40 686 346 352 31.0
5 Brazil 2010-07-21 6.07 22 891 33 348 55.0
6 Romania 2010-08-15 5.38 60 13 695 529 45.0
Slice rows by specific month:
In[]: time_df.loc[(time_df["datetime"].dt.month == 8)]
Out[]:
col_1 datetime col_3 col_4 col_5 col_6 col_7 col_8
2 Spain 2009-08-24 1.90 66 650 82 354 51.0
4 South Africa 2010-08-22 5.00 40 686 346 352 31.0
6 Romania 2010-08-15 5.38 60 13 695 529 45.0
9 India 2011-08-25 3.92 65 719 887 61 12.0
Slice rows by specific day:
In[]: time_df.loc[(time_df["datetime"].dt.day == 24)]
Out[]:
col_1 datetime col_3 col_4 col_5 col_6 col_7 col_8
2 Spain 2009-08-24 1.9 66 650 82 354 51.0
3 Rome 2010-07-24 2.0 76 408 84 538 98.0
Slice rows based on range between two dates:
'''
There are a few different methods to accomplish filtering rows by date.
Here we slice all the rows with dates greater than 2009-1-1 and less than 2010-8-1.
'''
#Step 1: create a date mask variable object
In[]: date_mask = (time_df['datetime'] > '2009-01-01') & (time_df['datetime'] <= '2010-8-1')
#Step 2: assign a new variable object dataframe to a .loc slice of the dataframe
In[]: new_datetime_df = time_df.loc[date_mask]
In[]: new_datetime_df
Out[]:
col_1 datetime col_3 col_4 col_5 col_6 col_7 col_8
0 Italy 2009-01-20 3.38 16 461 969 260 30.0
1 Greece 2009-02-23 7.00 42 770 107 229 85.0
2 Spain 2009-08-24 1.90 66 650 82 354 51.0
3 Rome 2010-07-24 2.00 76 408 84 538 98.0
5 Brazil 2010-07-21 6.07 22 891 33 348 55.0
Convert the date time column to year, month, and day columns:
'''
On occasion this can allow for more flexibility but is not really necessary.
'''
In[]: time_df['year'] = time_df['datetime'].dt.year
In[]: time_df['month'] = time_df['datetime'].dt.month
In[]: time_df['day'] = time_df['datetime'].dt.day
In[]: time_df
Out[]:
col_1 datetime col_3 col_4 col_5 col_6 col_7 col_8 year month day
0 Italy 2009-01-20 3.38 16 461 969 260 30.0 2009 1 20
1 Greece 2009-02-23 7.00 42 770 107 229 85.0 2009 2 23
2 Spain 2009-08-24 1.90 66 650 82 354 51.0 2009 8 24
3 Rome 2010-07-24 2.00 76 408 84 538 98.0 2010 7 24
4 South Africa 2010-08-22 5.00 40 686 346 352 31.0 2010 8 22
5 Brazil 2010-07-21 6.07 22 891 33 348 55.0 2010 7 21
6 Romania 2010-08-15 5.38 60 13 695 529 45.0 2010 8 15
7 Japan 2011-04-22 5.69 1 159 807 77 9.0 2011 4 22
8 Canada 2011-02-22 7.38 39 797 633 14 65.0 2011 2 22
9 India 2011-08-25 3.92 65 719 887 61 12.0 2011 8 25
'''
Also, convert the separate year, month, and day columns back to one datetime col.
'''
In[]: time_df['Date'] = pd.to_datetime(time_df[['year', 'month', 'day']])
In[]: time_df
Out[]:
col_6 col_7 col_8 year month day Date
0 ... 969 260 30.0 2009 1 20 2009-01-20
1 ... 107 229 85.0 2009 2 23 2009-02-23
2 ... 82 354 51.0 2009 8 24 2009-08-24
3 ... 84 538 98.0 2010 7 24 2010-07-24
4 ... 346 352 31.0 2010 8 22 2010-08-22
5 ... 33 348 55.0 2010 7 21 2010-07-21
6 ... 695 529 45.0 2010 8 15 2010-08-15
7 ... 807 77 9.0 2011 4 22 2011-04-22
8 ... 633 14 65.0 2011 2 22 2011-02-22
9 ... 887 61 12.0 2011 8 25 2011-08-25
Basic Statistics
Here I will use a new data frame to show some basic descriptive statistical methods.
I go into more advanced statistics in the data project examples (POST LINK HERE).
Here is our data:
'''
This data has no signifcant meaning outside of showing basic descriptive statistics
'''
In[]: stat_df
Out[]:
col_1 col_3 col_4 col_5 col_6 col_7 col_8
0 10 14 24 45 53 86 150
1 4 18 29 44 61 81 121
2 9 10 29 46 50 81 106
3 9 19 25 50 60 80 104
4 10 8 36 41 54 84 108
5 7 15 22 44 57 81 138
6 6 18 32 48 60 90 118
7 2 12 39 43 69 85 103
8 4 17 39 47 70 83 150
9 0 8 24 47 69 87 138
10 5 13 32 50 63 81 132
11 8 19 40 44 66 83 131
12 8 13 40 47 68 90 125
13 2 13 23 50 52 85 101
14 2 11 30 46 61 80 108
15 2 17 23 49 52 81 122
16 8 10 30 48 65 81 122
17 0 8 35 44 60 83 126
18 6 8 38 42 63 84 138
19 10 8 24 49 52 85 123
20 1 13 33 44 53 87 148
21 6 6 21 41 68 83 137
22 3 6 20 42 61 88 102
23 10 15 37 50 54 89 143
24 0 18 38 45 64 80 150
25 6 12 26 40 61 83 107
26 5 14 21 40 53 86 135
27 8 19 20 49 63 81 109
28 1 13 31 49 53 89 138
29 6 7 27 45 68 82 109
Return basic descriptive stats of each column:
'''
This method is a very simple built-in function in pandas.
'''
In[]: stat_df.describe()
Out[]:
col_1 col_3 col_4 col_5 col_6 col_7 col_8
count 30.000000 30.000000 30.000000 30.000000 30.00000 30.000000 30.000000
mean 5.266667 12.733333 29.600000 45.633333 60.10000 83.966667 124.733333
std 3.341828 4.176563 6.688126 3.178411 6.24417 3.145696 16.213943
min 0.000000 6.000000 20.000000 40.000000 50.00000 80.000000 101.000000
25% 2.000000 8.500000 24.000000 44.000000 53.25000 81.000000 108.250000
50% 6.000000 13.000000 29.500000 45.500000 61.00000 83.000000 124.000000
75% 8.000000 16.500000 35.750000 48.750000 64.75000 86.000000 138.000000
max 10.000000 19.000000 40.000000 50.000000 70.00000 90.000000 150.000000
Get the count of each value in a column:
'''
This also works for string data. This displays as:
<value> <count of that value>
'''
In[]: stat_df.col_1.value_counts()
Out[]:
6 5
10 4
2 4
8 4
0 3
4 2
9 2
5 2
1 2
7 1
3 1
Name: col_1, dtype: int64